Basic example#
Let’s use EOReader for the first time.
Imports#
import os
import tempfile
# EOReader
from eoreader.reader import Reader
from eoreader.bands import GREEN, NDVI, CLOUDS
Data#
First of all, we need some satellite data.
Let’s open a Sentinel-2 product.
path = os.path.join("/home", "data", "DATA", "PRODS", "S2", "PB 02.07+", "S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T094749.SAFE")
Create the Reader#
First, create the Reader object.
This object will automatically detect the type of constellation of your product.
It relies on the internal composition of the product (usually the presence of the metadata file), so please do not remove them.
The Reader is a singleton that should be called only once.
It can be used several times to open all your satellite products.
No need to extract the product here: archived Sentinel-2 are handled by EOReader.
reader = Reader()
Open the product#
The reader is used to open your product, just call the open function.
prod = reader.open(path)
prod
eoreader.S2Product 'S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T094749'
Attributes:
condensed_name: 20200114T065229_S2_T40REQ_L2A_094749
path: /home/data/DATA/PRODS/S2/PB 02.07+/S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T094749.SAFE
constellation: Sentinel-2
sensor type: Optical
product type: MSIL2A
default pixel size: 10.0
default resolution: 10.0
acquisition datetime: 2020-01-14T06:52:29
band mapping:
COASTAL_AEROSOL: 01
BLUE: 02
GREEN: 03
RED: 04
VEGETATION_RED_EDGE_1: 05
VEGETATION_RED_EDGE_2: 06
VEGETATION_RED_EDGE_3: 07
NIR: 08
NARROW_NIR: 8A
WATER_VAPOUR: 09
SWIR_1: 11
SWIR_2: 12
needs extraction: False
cloud cover: 63.86422
tile name: T40REQ
You can specify an output after creation, or on creation.
By default, your temporary files such as spectral bands are written on disk in this output.
You can prevent them from being written by specifying the remove_tmp argument.
prod = reader.open(path, output_path="/my/output", remove_tmp=True)
You can leverage the condensed_name for naming your output.
The condensed_name of your product is a shortened name with a pattern common to every EOReader product.
tmp_folder = tempfile.TemporaryDirectory()
prod.output = os.path.join(tmp_folder.name, prod.condensed_name)
Load#
Just load easily some bands and index. The load function outputs a dictionary of xarray.DataArray.
The bands can be called by their ID, name or mapped name.
For example, for Sentinel-3 OLCI you can use 7, Oa07 or YELLOW. For Landsat-8, you can use BLUE or 2.
# Load those bands as a xarray.Dataset
band_ds = prod.load([GREEN, NDVI, CLOUDS])
band_ds
<xarray.Dataset>
Dimensions: (band: 1, x: 10980, y: 10980)
Coordinates:
* band (band) int64 1
spatial_ref int64 0
* x (x) float64 5e+05 5e+05 ... 6.098e+05 6.098e+05
* y (y) float64 3e+06 3e+06 3e+06 ... 2.89e+06 2.89e+06
Data variables:
SpectralBandNames.GREEN (band, y, x) float32 0.1034 0.1134 ... nan nan
NDVI (band, y, x) float32 0.2816 0.2359 ... nan nan
CloudsBandNames.CLOUDS (band, y, x) float32 0.0 0.0 0.0 ... nan nan nan
Attributes:
long_name: GREEN NDVI CLOUDS
constellation: Sentinel-2
constellation_id: S2
product_path: /home/data/DATA/PRODS/S2/PB 02.07+/S2B_MSIL2A_20200114...
product_name: S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T...
product_filename: S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T...
instrument: MSI
product_type: MSIL2A
acquisition_date: 20200114T065229
condensed_name: 20200114T065229_S2_T40REQ_L2A_094749
orbit_direction: DESCENDING
cloud_cover: 63.86422# Load individual bands (as xarray.DataArray)
green = band_ds[GREEN]
ndvi = band_ds[NDVI]
clouds = band_ds[CLOUDS]
green
<xarray.DataArray <SpectralBandNames.GREEN: 'GREEN'> (band: 1, y: 10980,
x: 10980)>
array([[[0.1034, 0.1134, 0.1064, ..., nan, nan, nan],
[0.0969, 0.0895, 0.1094, ..., nan, nan, nan],
[0.1132, 0.0959, 0.1082, ..., nan, nan, nan],
...,
[1.488 , 1.4416, 1.3888, ..., nan, nan, nan],
[1.468 , 1.4472, 1.4072, ..., nan, nan, nan],
[1.4168, 1.4072, 1.4 , ..., nan, nan, nan]]],
dtype=float32)
Coordinates:
* band (band) int64 1
spatial_ref int64 0
* x (x) float64 5e+05 5e+05 5e+05 ... 6.098e+05 6.098e+05 6.098e+05
* y (y) float64 3e+06 3e+06 3e+06 ... 2.89e+06 2.89e+06 2.89e+06
Attributes: (12/13)
long_name: GREEN
constellation: Sentinel-2
constellation_id: S2
product_path: /home/data/DATA/PRODS/S2/PB 02.07+/S2B_MSIL2A_20200114...
product_name: S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T...
product_filename: S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T...
... ...
product_type: MSIL2A
acquisition_date: 20200114T065229
condensed_name: 20200114T065229_S2_T40REQ_L2A_094749
orbit_direction: DESCENDING
radiometry: reflectance
cloud_cover: 63.86422# Let's see what's inside your output folder
[f.relative_to(prod.output) for f in prod.output.glob("**/*.tif")]
[PosixPath('tmp_20200114T065229_S2_T40REQ_L2A_094749/20200114T065229_S2_T40REQ_L2A_094749_RED_10-00m_nodata.tif'),
PosixPath('tmp_20200114T065229_S2_T40REQ_L2A_094749/20200114T065229_S2_T40REQ_L2A_094749_NIR_10-00m_nodata.tif'),
PosixPath('tmp_20200114T065229_S2_T40REQ_L2A_094749/20200114T065229_S2_T40REQ_L2A_094749_NDVI_10-00m.tif'),
PosixPath('tmp_20200114T065229_S2_T40REQ_L2A_094749/20200114T065229_S2_T40REQ_L2A_094749_GREEN_10-00m_nodata.tif'),
PosixPath('tmp_20200114T065229_S2_T40REQ_L2A_094749/20200114T065229_S2_T40REQ_L2A_094749_CLOUDS_10-00m.tif')]
# Plot a subsampled version
green[:, ::10, ::10].plot()
<matplotlib.collections.QuadMesh at 0x7f6088eac050>
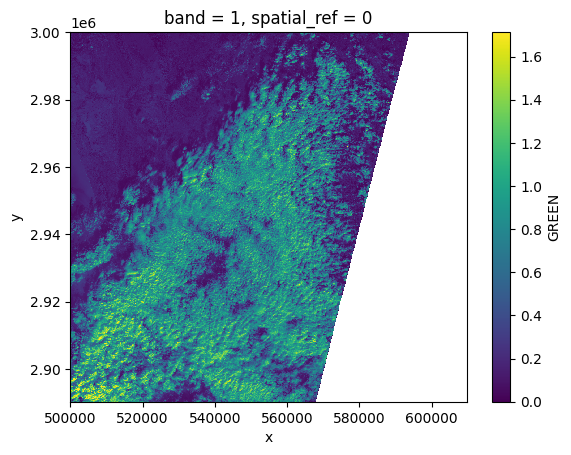
# Plot a subsampled version
ndvi[:, ::10, ::10].plot()
<matplotlib.collections.QuadMesh at 0x7f6088e54910>
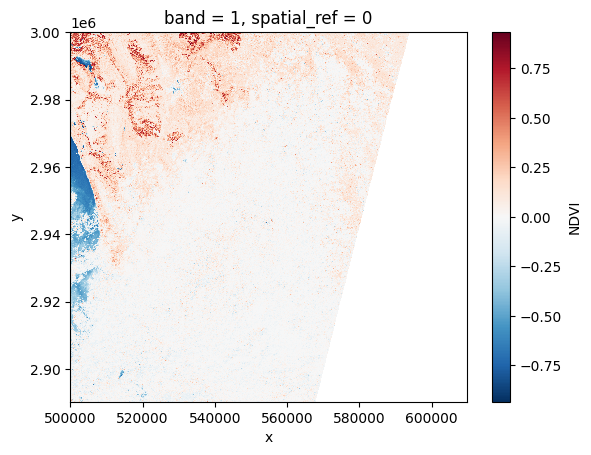
# Plot a subsampled version
clouds[:, ::10, ::10].plot()
<matplotlib.collections.QuadMesh at 0x7f6088e16810>
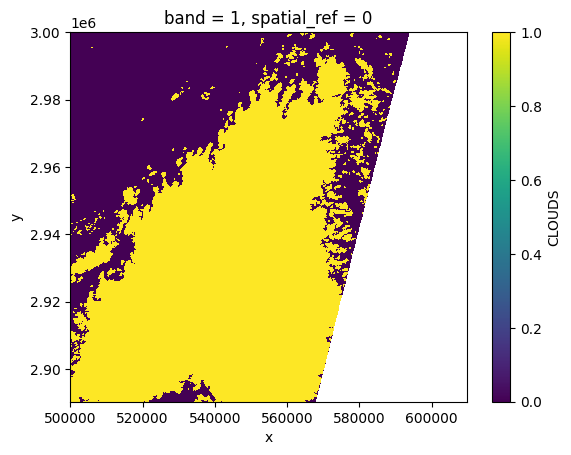
# Remove the output (it would have been best to call the temporary directory in a context manager)
tmp_folder.cleanup()
