DEM management#
DEM are used in two ways in EOReader:
for computing DEM bands (
DEM,SLOPE,HILLSHADE)for orthorectification (SAR, with SNAP or VHR data)
There are several things you need to know to handle DEMs properly.
# Imports
import os
import tempenv
import matplotlib.pyplot as plt
from eoreader.reader import Reader
from eoreader.bands import DEM, SLOPE, HILLSHADE, VV, RED
from eoreader.env_vars import DEM_PATH, SNAP_DEM_NAME
from eoreader.keywords import DEM_KW, SLOPE_KW, HILLSHADE_KW
from eoreader.products import SnapDems
# Create logger
import logging
from sertit import logs
logger = logging.getLogger("eoreader")
logs.init_logger(logger)
# Open the product
path = os.path.join("/home", "data", "DATA", "PRODS", "S2", "PB 02.07+", "S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T094749.SAFE")
prod = Reader().open(path)
# DEM paths
dem_folder = os.path.join("/home", "data", "DS2", "BASES_DE_DONNEES", "GLOBAL")
dtm = os.path.join(dem_folder, "MERIT_Hydrologically_Adjusted_Elevations", "MERIT_DEM.vrt")
dem = os.path.join(dem_folder, "COPDEM_30m", "COPDEM_30m.vrt")
dem_tif = os.path.join(dem_folder, "EUDEM_v2", "eudem_wgs84.tif")
DEM bands#
DEM bands are computed using the DEM stored in the EOREADER_DEM_PATH environment variable.
Without anything specified, DEM bands cannot be computed and an error will be thrown.
Note: SAR products cannot load a HILLSHADE band !
# Error when computing the DEM band: Iy hasn't been specified it
try:
prod.load(DEM)[DEM]
except ValueError as msg:
logger.error(msg)
2023-11-02 16:21:27,385 - [ERROR] - DEM path not set, unable to compute DEM bands! Please set the environment variable EOREADER_DEM_PATH or a DEM keyword.
# This works now
with tempenv.TemporaryEnvironment({DEM_PATH: dem}):
prod.load(DEM)[DEM]
2023-11-02 16:21:27,395 - [DEBUG] - Loading DEM bands ['DEM']
2023-11-02 16:21:27,395 - [DEBUG] - Warping DEM for 20200114T065229_S2_T40REQ_L2A_094749
2023-11-02 16:21:27,398 - [DEBUG] - Using DEM: /home/data/DS2/BASES_DE_DONNEES/GLOBAL/COPDEM_30m/COPDEM_30m.vrt
However, in some cases, you may want to specify a specific DEM to load a band from it.
The most common use of this is:
I want to compute the hillshade from a DEM
I want to compute the slope of a DTM as I don’t want the slopes of buildings or trees !
I want those two bands coregistered by default
When specifying a DEM in the load or stack function, you don’t need to set the EOREADER_DEM_PATH environment variable
dem_bands = prod.load(
[DEM, SLOPE, HILLSHADE],
**{
DEM_KW: dem,
SLOPE_KW: dtm, # We want a DTM here
HILLSHADE_KW: dem,
}
)
2023-11-02 16:21:28,644 - [DEBUG] - Loading DEM bands ['DEM', 'SLOPE', 'HILLSHADE']
2023-11-02 16:21:28,645 - [DEBUG] - Already existing DEM for 20200114T065229_S2_T40REQ_L2A_094749. Skipping process.
2023-11-02 16:21:28,937 - [DEBUG] - Warping DEM for 20200114T065229_S2_T40REQ_L2A_094749
2023-11-02 16:21:28,941 - [DEBUG] - Using DEM: /home/data/DS2/BASES_DE_DONNEES/GLOBAL/MERIT_Hydrologically_Adjusted_Elevations/MERIT_DEM.vrt
2023-11-02 16:21:28,997 - [DEBUG] - Computing slope for 20200114T065229_S2_T40REQ_L2A_094749
2023-11-02 16:21:39,609 - [DEBUG] - Already existing DEM for 20200114T065229_S2_T40REQ_L2A_094749. Skipping process.
2023-11-02 16:21:39,610 - [DEBUG] - Computing hillshade DEM for S2B_MSIL2A_20200114T065229_N0213_R020_T40REQ_20200114T094749
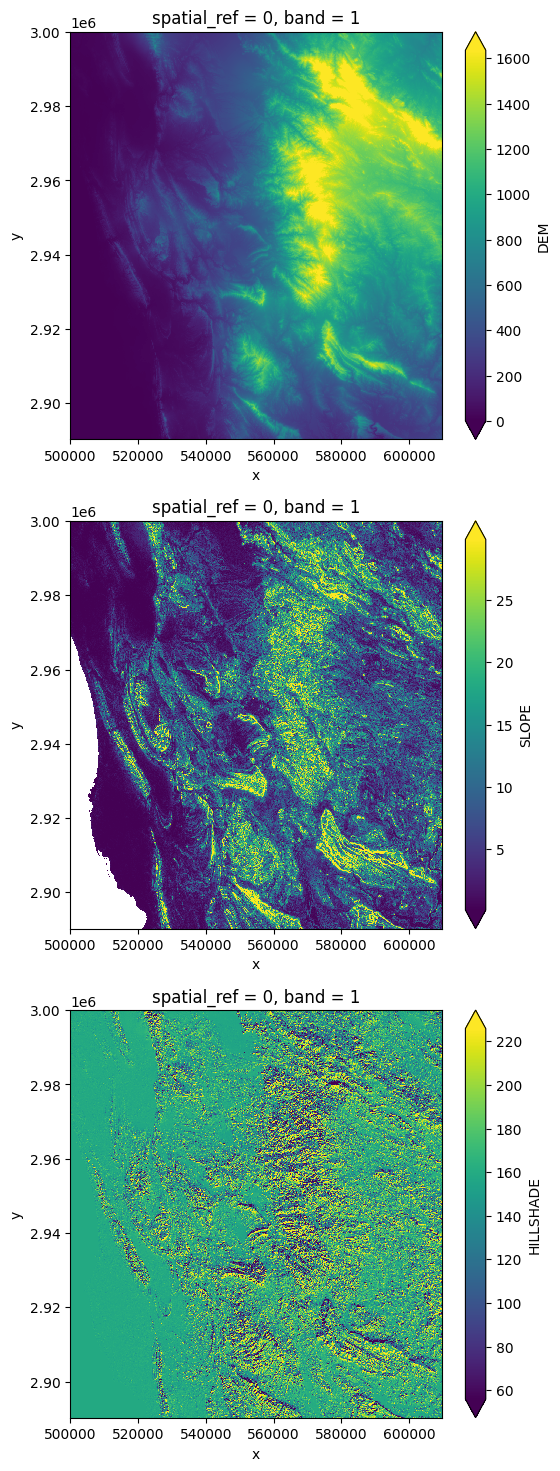
# Plot DEM bands
ncols = len(dem_bands)
plt.figure(figsize=(6, 6 * ncols))
i = 0
for key in dem_bands.keys():
axes = plt.subplot(3, 1, i+1)
dem_bands[key][0, ::10, ::10].plot.imshow(robust=True)
i += 1
Orthorectification#
SAR orthorectification (with SNAP)#
SNAP uses its own DEM mecanics: it downloads DEM tiles on its own and everything is managed without any intervention of the user.
By default, EOReader forces SNAP to use the COPDEM-30 (GLO-30) as its a global DTM.
However, this can be modified by setting the environment variable overriding the default DEM name used in SNAP.
The user can choose GETASSE30, SRTM 3Sec, External DEM…
If EOREADER_SNAP_DEM_NAME is set to External DEM, SNAP will use the DEM stored in EOREADER_DEM_PATH as an external DEM.
Warning: It seems that SNAP can only DEM with TIF format and not VRT.
Using such DEM format would result with the following error message: Error: No product reader found for /path/to/dem.vrt
# Open a SAR product
path = os.path.join("/home", "data", "DATA", "PRODS", "S1", "S1B_IW_GRDH_1SDV_20191215T180300_20191215T180325_019379_0249B2_C99C.SAFE")
prod = Reader().open(path)
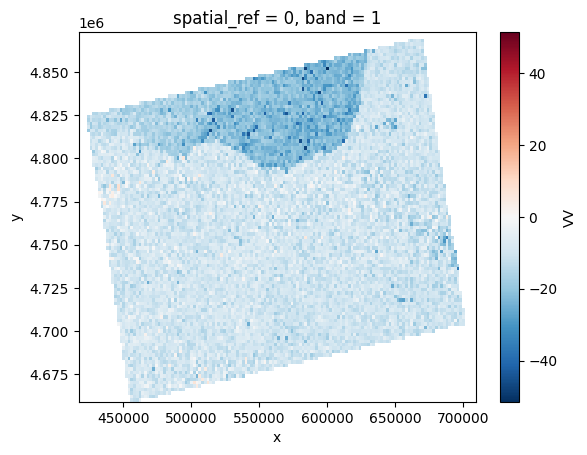
# Orthorectifying with COPDEM-30
vv = prod.load(VV, pixel_size=prod.pixel_size*100)[VV]
vv.plot()
SNAP Release version 9.0.0
SNAP home: /opt/snap/bin/..
SNAP debug: null
SNAP log level: WARNING
Java home: /opt/snap/jre/jre
Java version: 1.8.0_242
Processors: 16
Max memory: 40.9 GB
Cache size: 23.0 GB
Tile parallelism: 14
Tile size: 512 x 512 pixels
To configure your gpt memory usage:
Edit snap/bin/gpt.vmoptions
To configure your gpt cache size and parallelism:
Edit .snap/etc/snap.properties or gpt -c ${cachesize-in-GB}G -q ${parallelism}
Executing processing graph
OpenSearch: https://scihub.copernicus.eu/gnss/search?q=platformname:Sentinel-1 AND platformnumber:B AND producttype:AUX_POEORB AND beginposition:[2019-12-01T00:00:000Z TO 2019-12-31T24:00:000Z]
OpenSearch: 31 total results on 1 pages.
OpenSearch: https://scihub.copernicus.eu/gnss/search?q=platformname:Sentinel-1 AND platformnumber:B AND producttype:AUX_POEORB AND beginposition:[2019-12-01T00:00:000Z TO 2019-12-31T24:00:000Z]
version = 3.1
10%20%30%40%50%60%70%.80%90%Copernicus_DSM_COG_10_N44_00_W005_00_DEM.tif
Copernicus_DSM_COG_10_N44_00_W004_00_DEM.tif
Copernicus_DSM_COG_10_N44_00_W003_00_DEM.tif
Copernicus_DSM_COG_10_N44_00_W002_00_DEM.tif
Copernicus_DSM_COG_10_N44_00_W001_00_DEM.tif
Copernicus_DSM_COG_10_N43_00_W001_00_DEM.tif
Copernicus_DSM_COG_10_N43_00_W005_00_DEM.tif
Copernicus_DSM_COG_10_N43_00_W004_00_DEM.tif
Copernicus_DSM_COG_10_N43_00_W003_00_DEM.tif
Copernicus_DSM_COG_10_N43_00_W002_00_DEM.tif
Copernicus_DSM_COG_10_N42_00_W001_00_DEM.tif
Copernicus_DSM_COG_10_N42_00_W002_00_DEM.tif
Copernicus_DSM_COG_10_N42_00_W005_00_DEM.tif
Copernicus_DSM_COG_10_N42_00_W004_00_DEM.tif
Copernicus_DSM_COG_10_N42_00_W003_00_DEM.tif
done.
2023-11-02 16:22:21,731 - [DEBUG] - Loading bands ['VV']
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 16:22:23,242 - [DEBUG] - Pre-process SAR image
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 16:25:07,822 - [DEBUG] - Converting DIMAP to GeoTiff
<matplotlib.collections.QuadMesh at 0x7f3c34423690>
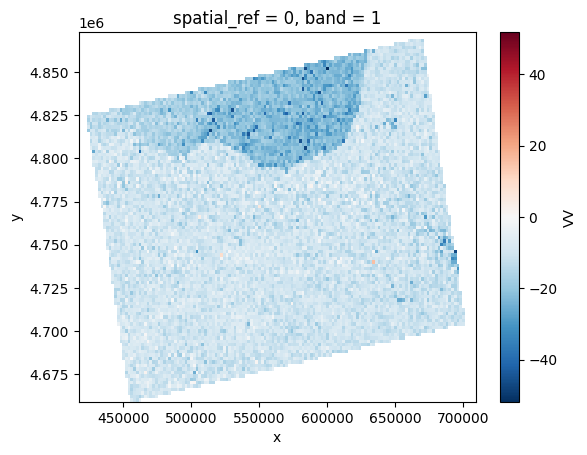
# Orthorectifying with GETASSE
prod.clean_tmp()
with tempenv.TemporaryEnvironment(
{
SNAP_DEM_NAME: SnapDems.GETASSE30.value
}
):
vv2 = prod.load(VV, pixel_size=prod.pixel_size*100)[VV]
vv2.plot()
SNAP Release version 9.0.0
SNAP home: /opt/snap/bin/..
SNAP debug: null
SNAP log level: WARNING
Java home: /opt/snap/jre/jre
Java version: 1.8.0_242
Processors: 16
Max memory: 40.9 GB
Cache size: 23.0 GB
Tile parallelism: 14
Tile size: 512 x 512 pixels
To configure your gpt memory usage:
Edit snap/bin/gpt.vmoptions
To configure your gpt cache size and parallelism:
Edit .snap/etc/snap.properties or gpt -c ${cachesize-in-GB}G -q ${parallelism}
Executing processing graph
version = 3.1
10%20%30%40%50%60%70%.80%90% done.
2023-11-02 16:25:08,118 - [DEBUG] - Loading bands ['VV']
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 16:25:09,395 - [DEBUG] - Pre-process SAR image
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 16:26:27,344 - [DEBUG] - Converting DIMAP to GeoTiff
# Orthorectifying with custom TIF DEM (EUDEM)
with tempenv.TemporaryEnvironment(
{
SNAP_DEM_NAME: SnapDems.EXT_DEM.value,
DEM_PATH: dem_tif
}
):
vv = prod.load(VV, pixel_size=prod.pixel_size*100)[VV]
VHR orthorectification#
Some products are not orthorectified by default (according what product type you bought).
It may be wanted, in order to control yourself the orthorectification.
EOReader can orthorectify VHR products: DIMAP (Pleiades and SPOT) and Maxar (GeoEye and WorldView) products.
Warning: The orthorectification may take a while !
# Open a VHR product in sensor geometry
path = os.path.join("/home", "data", "DATA", "PRODS", "PLEIADES", "3302499201", "IMG_PHR1A_MS_004")
prod = Reader().open(path)
# Error when orthorectifying the RED band: Iy hasn't been specified it
try:
prod.load(RED)[RED]
except ValueError as msg:
logger.error(msg)
2023-11-02 16:26:28,392 - [DEBUG] - Loading bands ['RED']
2023-11-02 16:26:28,393 - [INFO] - Manually orthorectified stack not given by the user. Reprojecting whole stack, this may take a while. (May be inaccurate on steep terrain, depending on the DEM pixel size)
2023-11-02 16:26:28,393 - [ERROR] - As you are using a non orthorectified VHR product (/home/data/DATA/PRODS/PLEIADES/3302499201/IMG_PHR1A_MS_004), you must provide a valid DEM through the EOREADER_DEM_PATH environment variable
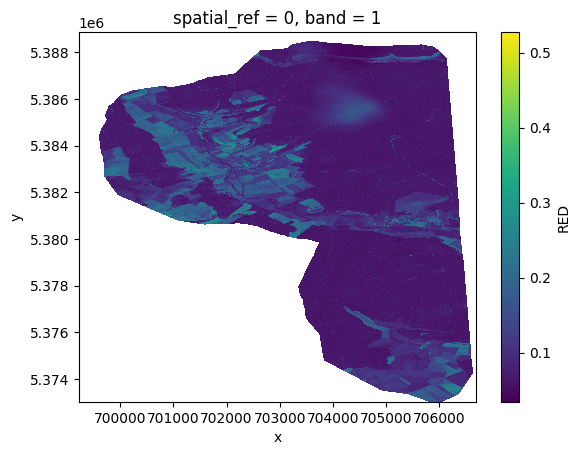
# With environment variable
with tempenv.TemporaryEnvironment({DEM_PATH: dem}):
red = prod.load(RED)[RED]
red.plot()
2023-11-02 16:26:28,400 - [DEBUG] - Loading bands ['RED']
2023-11-02 16:26:28,400 - [INFO] - Manually orthorectified stack not given by the user. Reprojecting whole stack, this may take a while. (May be inaccurate on steep terrain, depending on the DEM pixel size)
2023-11-02 16:26:29,676 - [DEBUG] - Orthorectifying data with /home/data/DS2/BASES_DE_DONNEES/GLOBAL/COPDEM_30m/COPDEM_30m.vrt
2023-11-02 16:28:10,147 - [DEBUG] - Read RED
2023-11-02 16:28:10,167 - [DEBUG] - Manage nodata for band RED
ERROR 1: Cannot import urn:ogc:def:derivedCRSType:OGC:1.0:image due to ALLOW_FILE_ACCESS=NO
2023-11-02 16:28:10,287 - [INFO] - Orthorectifying ROI
2023-11-02 16:28:10,322 - [DEBUG] - Rasterizing ROI
2023-11-02 16:28:10,353 - [DEBUG] - Reprojecting ROI
2023-11-02 16:28:10,354 - [DEBUG] - Orthorectifying data with /home/data/DS2/BASES_DE_DONNEES/GLOBAL/COPDEM_30m/COPDEM_30m.vrt
2023-11-02 16:29:41,274 - [DEBUG] - Revectorizing ROI
2023-11-02 16:29:42,188 - [DEBUG] - Converting RED to reflectance
<matplotlib.collections.QuadMesh at 0x7f3c2c663250>
# With keyword
red = prod.load(RED, **{DEM_KW: dem})[RED]
