SAR example#
Let’s use EOReader with SAR data.
Warning: SAR data is processed with SNAP, so be sure to have it installed and that GPT is in your path.
Imports#
import os
import matplotlib.pyplot as plt
# EOReader
from eoreader.reader import Reader
from eoreader.bands import VV, HH, VV_DSPK, HH_DSPK, HILLSHADE, SLOPE, to_str
from eoreader.env_vars import DEM_PATH
Create logger#
# Create logger
import logging
from sertit import logs
logger = logging.getLogger("eoreader")
logs.init_logger(logger)
Open the COSMO-SkyMed product#
Please be aware that:
EOReader will orthorectify your SAR data to get UTM tiles.
complex data is not handled as is, EOReader will convert them to ground range.
# First of all, we need some VHR data, let's use some COSMO-SkyMed data
path = os.path.join("/home", "data", "DATA", "PRODS", "COSMO", "1st_GEN", "1001512-735097")
# Open your product
prod = Reader().open(path, remove_tmp=True)
prod
eoreader.CskProduct 'CSKS4_DGM_B_HI_09_HH_RA_FF_20201008224018_20201008224025'
Attributes:
condensed_name: 20201008T224018_CSK_HH_HI_DGM
path: /home/data/DATA/PRODS/COSMO/1st_GEN/1001512-735097
constellation: COSMO-SkyMed
sensor type: SAR
product type: DGM
default pixel size: 2.5
default resolution: 5.0
acquisition datetime: 2020-10-08T22:40:18.446381
band mapping:
HH: HH
HH_DSPK: HH_DSPK
needs extraction: True
orbit direction: ASCENDING
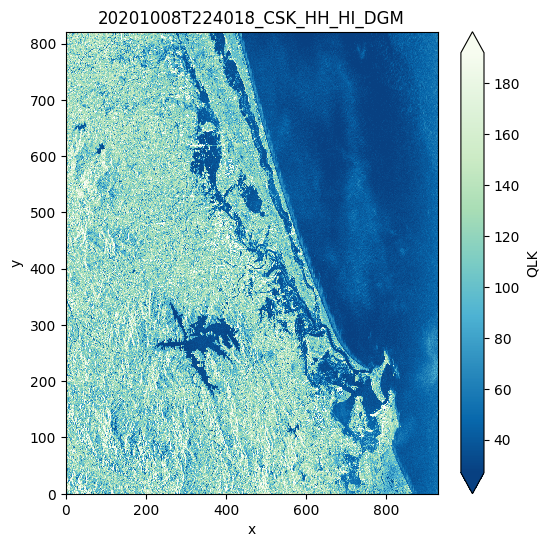
# Plot the quicklook
prod.plot()
/opt/conda/lib/python3.11/site-packages/rasterio/__init__.py:304: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
dataset = DatasetReader(path, driver=driver, sharing=sharing, **kwargs)
/opt/conda/lib/python3.11/site-packages/rasterio/__init__.py:304: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
dataset = DatasetReader(path, driver=driver, sharing=sharing, **kwargs)
/opt/conda/lib/python3.11/site-packages/rioxarray/_io.py:1132: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
warnings.warn(str(rio_warning.message), type(rio_warning.message)) # type: ignore
/opt/conda/lib/python3.11/site-packages/rasterio/__init__.py:314: NotGeoreferencedWarning: The given matrix is equal to Affine.identity or its flipped counterpart. GDAL may ignore this matrix and save no geotransform without raising an error. This behavior is somewhat driver-specific.
dataset = writer(
# Get the band information
prod.bands
eoreader.SarBand 'HH'
Attributes:
id: HH
eoreader_name: HH
gsd (m): 2.5
asset_role: intensity
eoreader.SarBand 'HH_DSPK'
Attributes:
id: HH_DSPK
eoreader_name: HH_DSPK
gsd (m): 2.5
asset_role: intensity
# Print some data
print(f"Acquisition datetime: {prod.datetime}")
print(f"Condensed name: {prod.condensed_name}")
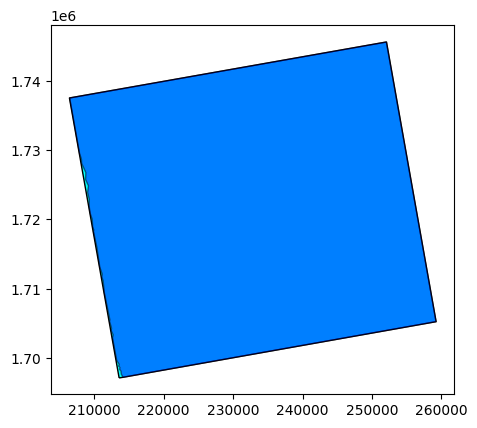
# Open here some more interesting geographical data: extent and footprint
extent = prod.extent()
footprint = prod.footprint()
base = extent.plot(color='cyan', edgecolor='black')
footprint.plot(ax=base, color='blue', edgecolor='black', alpha=0.5)
Acquisition datetime: 2020-10-08 22:40:18.446381
Condensed name: 20201008T224018_CSK_HH_HI_DGM
SNAP Release version 9.0.0
SNAP home: /opt/snap/bin/..
SNAP debug: null
SNAP log level: WARNING
Java home: /opt/snap/jre/jre
Java version: 1.8.0_242
Processors: 16
Max memory: 40.9 GB
Cache size: 23.0 GB
Tile parallelism: 14
Tile size: 512 x 512 pixels
To configure your gpt memory usage:
Edit snap/bin/gpt.vmoptions
To configure your gpt cache size and parallelism:
Edit .snap/etc/snap.properties or gpt -c ${cachesize-in-GB}G -q ${parallelism}
Executing processing graph
Copernicus_DSM_COG_10_N15_00_E108_00_DEM.tif
....10%....20%....30%....40%....50%....60%....70%....80%....90% done.
/opt/conda/lib/python3.11/site-packages/rasterio/__init__.py:304: NotGeoreferencedWarning: Dataset has no geotransform, gcps, or rpcs. The identity matrix will be returned.
dataset = DatasetReader(path, driver=driver, sharing=sharing, **kwargs)
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 15:51:00,059 - [DEBUG] - Pre-process SAR image
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 16:07:11,798 - [DEBUG] - Converting DIMAP to GeoTiff
<Axes: >
For SAR data, the footprint needs the orthorectified data !
For that, SNAP uses its own DEM, but you can change it when positionning the EOREADER_SNAP_DEM_NAME environment variable.
Available DEMs are:
ACE2_5MinACE30ASTER 1sec GDEMCopernicus 30m Global DEMCopernicus 90m Global DEMGETASSE30SRTM 1Sec HGTSRTM 3SecExternal DEM
Warning:
If External DEM is set, you must specify the DEM you want by positioning the EOREADER_DEM_PATH to a DEM that can be read by SNAP.
Load bands#
# Set the DEM
os.environ[DEM_PATH] = os.path.join("/home", "data", "DS2", "BASES_DE_DONNEES", "GLOBAL", "COPDEM_30m", "COPDEM_30m.vrt")
# Select some bands you wish to load without knowing if they exist
bands = [VV, HH, VV_DSPK, HH_DSPK, HILLSHADE, SLOPE]
# Only keep those selected
ok_bands = [band for band in bands if prod.has_band(band)]
# This product does not have VV band and HILLSHADE band cannot be computed from SAR band
print(to_str(ok_bands))
['HH', 'HH_DSPK', 'SLOPE']
# Load those bands as a xarray.Dataset, with a 20m pixel size
band_ds = prod.load(ok_bands, pixel_size=20.)
band_ds[HH]
Executing processing graph
first_line_time metadata value is null
last_line_time metadata value is null
....10%....20%....30%....40%....50%....60%....70%....80%....90% done.
2023-11-02 16:07:30,658 - [DEBUG] - Loading bands ['HH', 'HH_DSPK']
2023-11-02 16:07:30,662 - [DEBUG] - Despeckling HH
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s2tbx [reason: /opt/snap/s2tbx]
SEVERE: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Failed to scan /opt/snap/s3tbx [reason: /opt/snap/s3tbx]
2023-11-02 16:08:49,827 - [DEBUG] - Loading DEM bands ['SLOPE']
2023-11-02 16:08:49,828 - [DEBUG] - Warping DEM for 20201008T224018_CSK_HH_HI_DGM
2023-11-02 16:08:49,831 - [DEBUG] - Using DEM: /home/data/DS2/BASES_DE_DONNEES/GLOBAL/COPDEM_30m/COPDEM_30m.vrt
2023-11-02 16:08:50,653 - [DEBUG] - Computing slope for 20201008T224018_CSK_HH_HI_DGM
<xarray.DataArray <SarBandNames.HH: 'HH'> (band: 1, y: 2473, x: 2688)>
array([[[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
Coordinates:
* x (x) float64 2.058e+05 2.059e+05 ... 2.596e+05 2.596e+05
* y (y) float64 1.746e+06 1.746e+06 ... 1.697e+06 1.697e+06
spatial_ref int64 0
* band (band) int64 1
Attributes:
long_name: HH
constellation: COSMO-SkyMed
constellation_id: CSK
product_path: /home/data/DATA/PRODS/COSMO/1st_GEN/1001512-735097
product_name: CSKS4_DGM_B_HI_09_HH_RA_FF_20201008224018_20201008224025
product_filename: 1001512-735097
instrument: SAR-2000
product_type: DGM
acquisition_date: 20201008T224018
condensed_name: 20201008T224018_CSK_HH_HI_DGM
orbit_direction: ASCENDINGThis can lead the Terrain Correction step to create large nodata area when projecting on a DEM.
If it happens, you can set the keyword SAR_INTERP_NA to True when loading or stacking SAR data to fill these area with interpolated data.
from eoreader.keywords import SAR_INTERP_NA
band_dict = prod.load(
ok_bands,
pixel_size=20.,
**{SAR_INTERP_NA: True}
)
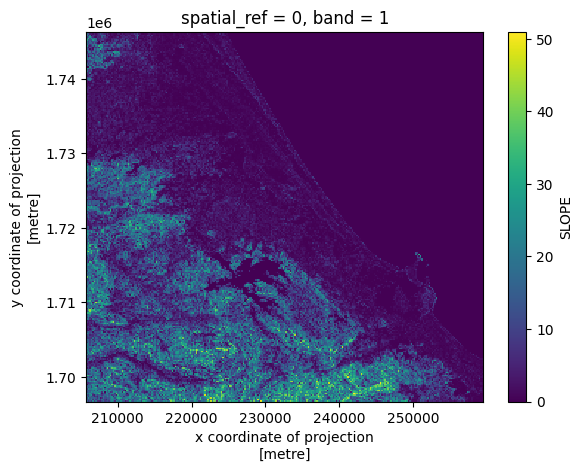
# Plot a subsampled version
band_ds[SLOPE][:, ::10, ::10].plot()
<matplotlib.collections.QuadMesh at 0x7f59f0193450>
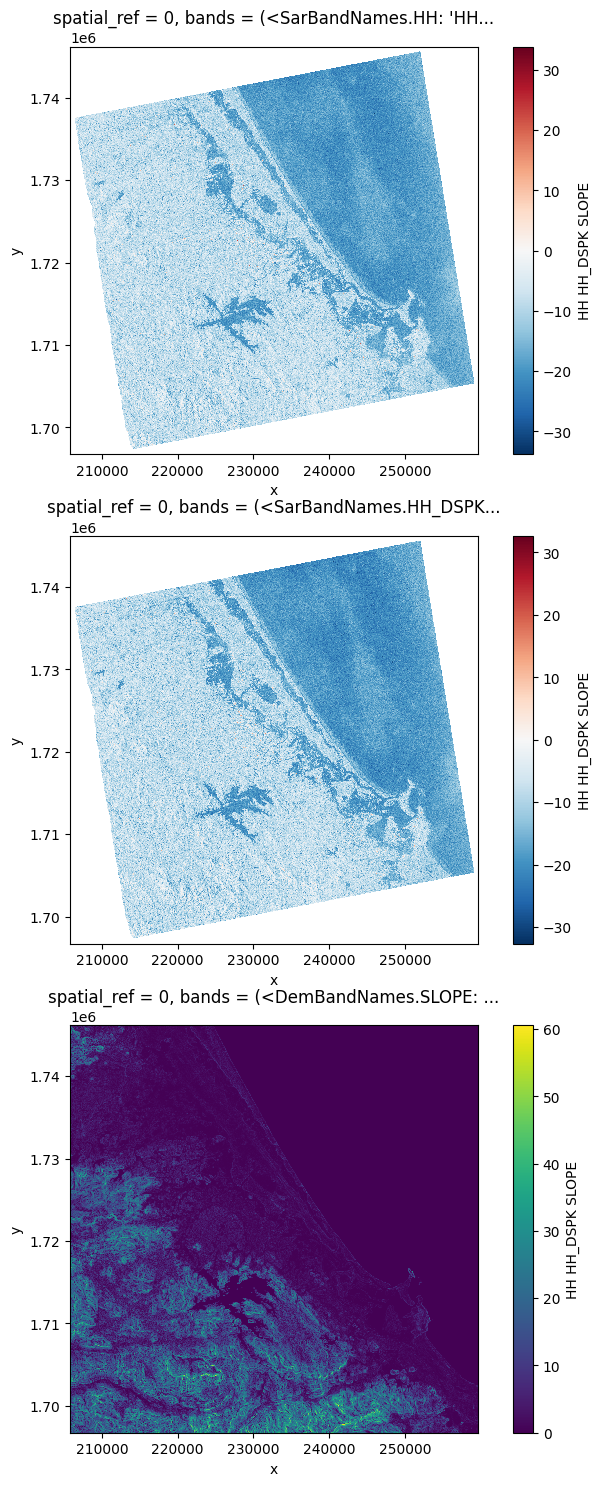
Stack some data#
# You can also stack those bands
stack = prod.stack(ok_bands)
stack
2023-11-02 16:08:53,400 - [DEBUG] - Loading bands ['HH', 'HH_DSPK']
2023-11-02 16:08:53,435 - [DEBUG] - Loading DEM bands ['SLOPE']
2023-11-02 16:08:53,436 - [DEBUG] - Already existing DEM for 20201008T224018_CSK_HH_HI_DGM. Skipping process.
2023-11-02 16:08:53,437 - [DEBUG] - Already existing slope DEM for 20201008T224018_CSK_HH_HI_DGM. Skipping process.
2023-11-02 16:09:14,902 - [DEBUG] - Stacking
/opt/conda/lib/python3.11/site-packages/xarray/core/indexes.py:662: RuntimeWarning: '<' not supported between instances of 'SarBandNames' and 'SarBandNames', sort order is undefined for incomparable objects.
new_pd_index = pd_indexes[0].append(pd_indexes[1:])
<xarray.DataArray 'HH_HH_DSPK_SLOPE' (bands: 3, y: 19786, x: 21505)>
dask.array<transpose, shape=(3, 19786, 21505), dtype=float32, chunksize=(1, 2048, 2048), chunktype=numpy.ndarray>
Coordinates:
* x (x) float64 2.058e+05 2.058e+05 ... 2.596e+05 2.596e+05
* y (y) float64 1.746e+06 1.746e+06 ... 1.697e+06 1.697e+06
spatial_ref int64 0
* bands (bands) object MultiIndex
* variable (bands) object SarBandNames.HH ... DemBandNames.SLOPE
* band (bands) int64 1 1 1
Attributes:
long_name: HH HH_DSPK SLOPE
constellation: COSMO-SkyMed
constellation_id: CSK
product_path: /home/data/DATA/PRODS/COSMO/1st_GEN/1001512-735097
product_name: CSKS4_DGM_B_HI_09_HH_RA_FF_20201008224018_20201008224025
product_filename: 1001512-735097
instrument: SAR-2000
product_type: DGM
acquisition_date: 20201008T224018
condensed_name: 20201008T224018_CSK_HH_HI_DGM
orbit_direction: ASCENDING# Plot a subsampled version
nrows = len(stack)
fig, axes = plt.subplots(nrows=nrows, figsize=(3 * nrows, 6 * nrows), subplot_kw={"box_aspect": 1})
for i in range(nrows):
stack[i, ::10, ::10].plot(x="x", y="y", ax=axes[i])