Custom stacks
Contents
Custom stacks#
Let’s use EOReader with custom stacks.
# EOReader Imports
import os
import xarray as xr
from eoreader.reader import Reader
from eoreader.products import SensorType
from eoreader.bands import BLUE, GREEN, RED, NIR, SWIR_1, VV, VV_DSPK, SLOPE, HILLSHADE
from sertit import display
reader = Reader()
# Create logger
import logging
from sertit import logs
logs.init_logger(logging.getLogger("eoreader"))
# Set a DEM
from eoreader.env_vars import DEM_PATH
os.environ[DEM_PATH] = os.path.join("/home", "data", "DS2", "BASES_DE_DONNEES", "GLOBAL", "COPDEM_30m",
"COPDEM_30m.vrt")
Custom stack with minimum data#
For both SAR and optical stacks, the two minimum keywords to provide are:
band_map: a dictionary mapping the satellite band to the band number (starting to 1, in GDAL style)sensor_type: EitherSARorOPTICAL(a string or a SensorType Enum)
# Paths
stack_folder = os.path.join("/home", "data", "DS3", "CI", "eoreader", "others")
opt_path = os.path.join(stack_folder, "20200310T030415_WV02_Ortho_BGRN_STK.tif")
sar_path = os.path.join(stack_folder, "20210827T162210_ICEYE_SC_GRD_STK.tif")
# Optical minimum example
opt_prod = reader.open(opt_path,
custom=True,
sensor_type="OPTICAL", # With a string
band_map={BLUE: 1, GREEN: 2, RED: 3, NIR: 4, SWIR_1: 5})
opt_prod
eoreader.CustomProduct '20200310T030415_WV02_Ortho_BGRN_STK'
Attributes:
condensed_name: 20221010T141943_CUSTOM_CUSTOM
path: /home/data/DS3/CI/eoreader/others/20200310T030415_WV02_Ortho_BGRN_STK.tif
constellation: CUSTOM
sensor type: Optical
product type: CUSTOM
default resolution: 7.999924228754893
acquisition datetime: 2022-10-10T14:19:43.482589
band mapping:
BLUE: 1
GREEN: 2
RED: 3
NIR: 4
SWIR_1: 5
needs extraction: False
opt_stack = opt_prod.stack([BLUE, GREEN, RED])
2022-10-10 14:19:43,489 - [DEBUG] - Loading bands ['GREEN', 'BLUE', 'RED']
2022-10-10 14:19:44,913 - [DEBUG] - Stacking
2022-10-10 14:19:44,938 - [DEBUG] - Saving stack
xr.plot.imshow(opt_stack.copy(data=display.scale(opt_stack.data)))
<matplotlib.image.AxesImage at 0x7fe664643e50>
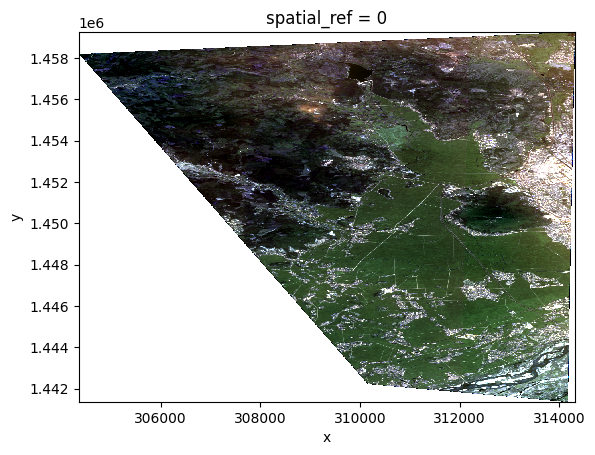
opt_stack
<xarray.DataArray 'BLUE_GREEN_RED' (z: 3, y: 2237, x: 1244)>
array([[[ nan, nan, nan, ..., 0.02729181,
0.03021449, 0.0321508 ],
[ nan, nan, nan, ..., 0.03289769,
0.03252383, 0.03231718],
[ nan, nan, nan, ..., 0.03253607,
0.03250813, 0.03260763],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[ nan, nan, nan, ..., 0.0325688 ,
0.03575394, 0.03786882],
[ nan, nan, nan, ..., 0.03874811,
0.0377332 , 0.0372853 ],
[ nan, nan, nan, ..., 0.03795209,
0.03785328, 0.03810363],
...
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[ nan, nan, nan, ..., 0.02202989,
0.02403895, 0.02508134],
[ nan, nan, nan, ..., 0.02564428,
0.02424301, 0.02346394],
[ nan, nan, nan, ..., 0.0244639 ,
0.02421321, 0.02448287],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]]], dtype=float32)
Coordinates:
* x (x) float64 3.044e+05 3.044e+05 ... 3.143e+05 3.143e+05
* y (y) float64 1.459e+06 1.459e+06 ... 1.441e+06 1.441e+06
spatial_ref int64 0
* z (z) object MultiIndex
* variable (z) object 'BLUE' 'GREEN' 'RED'
* band (z) int64 1 1 1
Attributes:
long_name: BLUE GREEN RED
constellation: CUSTOM
constellation_id: CUSTOM
product_path: /home/data/DS3/CI/eoreader/others/20200310T030415_WV02...
product_name: 20200310T030415_WV02_Ortho_BGRN_STK
product_filename: 20200310T030415_WV02_Ortho_BGRN_STK
instrument: CUSTOM
product_type: CUSTOM
acquisition_date: 20221010T141944
condensed_name: 20221010T141943_CUSTOM_CUSTOM
orbit_direction: None# SAR minimum example
sar_prod = reader.open(sar_path,
custom=True,
sensor_type=SensorType.SAR, # With the Enum
band_map={VV: 1, VV_DSPK: 2})
sar_prod
eoreader.CustomProduct '20210827T162210_ICEYE_SC_GRD_STK'
Attributes:
condensed_name: 20221010T141945_CUSTOM_CUSTOM
path: /home/data/DS3/CI/eoreader/others/20210827T162210_ICEYE_SC_GRD_STK.tif
constellation: CUSTOM
sensor type: SAR
product type: CUSTOM
default resolution: 47.995955510616774
acquisition datetime: 2022-10-10T14:19:45.926447
band mapping:
VV: 1
VV_DSPK: 2
needs extraction: False
sar_stack = sar_prod.stack([SLOPE, VV, VV_DSPK])
2022-10-10 14:19:45,934 - [DEBUG] - Loading bands ['VV_DSPK', 'VV']
2022-10-10 14:19:47,061 - [DEBUG] - Warping DEM for 20221010T141945_CUSTOM_CUSTOM
2022-10-10 14:19:47,064 - [DEBUG] - Using DEM: /home/data/DS2/BASES_DE_DONNEES/GLOBAL/COPDEM_30m/COPDEM_30m.vrt
2022-10-10 14:19:51,042 - [DEBUG] - Computing slope for 20221010T141945_CUSTOM_CUSTOM
2022-10-10 14:19:51,398 - [DEBUG] - Stacking
2022-10-10 14:19:51,437 - [DEBUG] - Saving stack
xr.plot.imshow(sar_stack.copy(data=display.scale(sar_stack.data)))
<matplotlib.image.AxesImage at 0x7fe6625eba00>

sar_stack
<xarray.DataArray 'SLOPE_VV_VV_DSPK' (z: 3, y: 2748, x: 2967)>
array([[[1.1417845 , 0.9661645 , 0.88848215, ..., 0. ,
0. , 0. ],
[0.91908467, 0.8988768 , 0.9166924 , ..., 0. ,
0. , 0. ],
[1.0019214 , 0.84933126, 0.86957526, ..., 0. ,
0. , 0. ],
...,
[0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[0. , 0. , 0. , ..., 0. ,
0. , 0. ],
[0. , 0. , 0. , ..., 0. ,
0. , 0. ]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]],
[[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan]]], dtype=float32)
Coordinates:
* x (x) float64 6.7e+05 6.701e+05 6.701e+05 ... 8.124e+05 8.124e+05
* y (y) float64 1.113e+04 1.109e+04 ... -1.206e+05 -1.207e+05
spatial_ref int64 0
* z (z) object MultiIndex
* variable (z) object 'SLOPE' 'VV' 'VV_DSPK'
* band (z) int64 1 1 1
Attributes:
long_name: SLOPE VV VV_DSPK
constellation: CUSTOM
constellation_id: CUSTOM
product_path: /home/data/DS3/CI/eoreader/others/20210827T162210_ICEY...
product_name: 20210827T162210_ICEYE_SC_GRD_STK
product_filename: 20210827T162210_ICEYE_SC_GRD_STK
instrument: CUSTOM
product_type: CUSTOM
acquisition_date: 20221010T141951
condensed_name: 20221010T141945_CUSTOM_CUSTOM
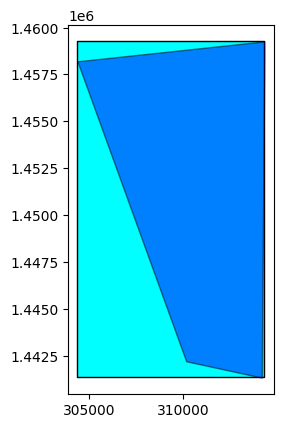
orbit_direction: None# You can compute the footprint and the extent
extent = opt_prod.extent()
footprint = opt_prod.footprint()
base = extent.plot(color='cyan', edgecolor='black')
footprint.plot(ax=base, color='blue', edgecolor='black', alpha=0.5)
<AxesSubplot: >
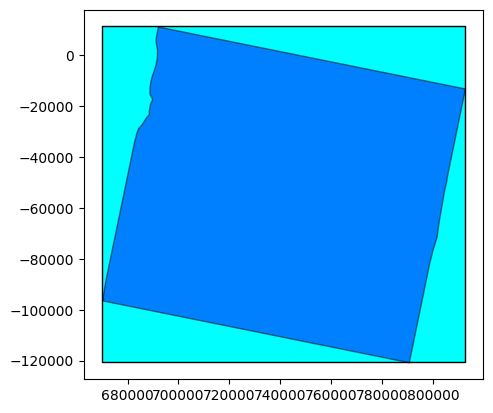
extent = sar_prod.extent()
footprint = sar_prod.footprint()
base = extent.plot(color='cyan', edgecolor='black')
footprint.plot(ax=base, color='blue', edgecolor='black', alpha=0.5)
<AxesSubplot: >
Custom stack with full data#
If you know them, it is best to give EOReader all the data you know about your stack:
name: product name. If not provided, the filename will be usedacquisition_datetime: product acquisition datetime. If not provided, the datetime of the creation of the object will be usedplatform: product platform. If not provided,CUSTOMwill be set. Either a string of a Platform enum.product_type: product type. If not provided,CUSTOMwill be set.default_resolution: product default resolution. If not provided, the stack resolution will be used.
For optical products, two additional keyword can be set to compute the hillshade band:
sun_azimuthsun_zenith
# Optical
opt_prod = reader.open(
opt_path,
custom=True,
name="20200310T030415_WV02_Ortho",
acquisition_datetime="20200310T030415",
sensor_type=SensorType.OPTICAL,
platform="WV02",
product_type="Ortho",
default_resolution=2.0,
sun_azimuth=10.0,
sun_zenith=20.0,
band_map={BLUE: 1, GREEN: 2, RED: 3, NIR: 4, SWIR_1: 5},
)
hillshade = opt_prod.load(HILLSHADE)[HILLSHADE]
2022-10-10 14:19:54,429 - [WARNING] - acquisition_datetime is not taken into account as it doesn't belong to the handled keys: ['name', 'sensor_type', 'datetime', 'band_map', 'constellation', 'instrument', 'resolution', 'product_type', 'sun_azimuth', 'sun_zenith', 'orbit_direction', 'cloud_cover']
2022-10-10 14:19:54,429 - [WARNING] - platform is not taken into account as it doesn't belong to the handled keys: ['name', 'sensor_type', 'datetime', 'band_map', 'constellation', 'instrument', 'resolution', 'product_type', 'sun_azimuth', 'sun_zenith', 'orbit_direction', 'cloud_cover']
2022-10-10 14:19:54,430 - [WARNING] - default_resolution is not taken into account as it doesn't belong to the handled keys: ['name', 'sensor_type', 'datetime', 'band_map', 'constellation', 'instrument', 'resolution', 'product_type', 'sun_azimuth', 'sun_zenith', 'orbit_direction', 'cloud_cover']
2022-10-10 14:19:54,446 - [DEBUG] - Warping DEM for 20221010T141954_CUSTOM_Ortho
2022-10-10 14:19:54,449 - [DEBUG] - Using DEM: /home/data/DS2/BASES_DE_DONNEES/GLOBAL/COPDEM_30m/COPDEM_30m.vrt
2022-10-10 14:19:55,091 - [DEBUG] - Computing hillshade DEM for 20200310T030415_WV02_Ortho
hillshade.plot()
<matplotlib.collections.QuadMesh at 0x7fe64c70cc40>
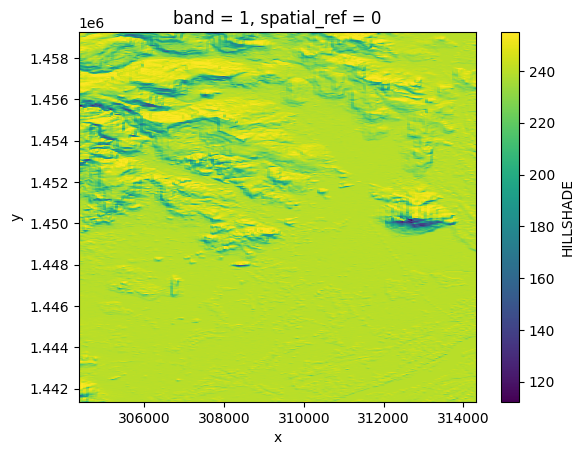
hillshade
<xarray.DataArray 'HILLSHADE' (band: 1, y: 2237, x: 1244)>
array([[[243.51416, 244.15453, 244.76277, ..., 239.27824, 239.38185,
239.48503],
[241.75093, 242.62413, 243.46097, ..., 239.40317, 239.44379,
239.4857 ],
[239.75554, 240.88219, 241.96497, ..., 239.65657, 239.58911,
239.52312],
...,
[247.7831 , 248.34468, 246.92708, ..., 239.21782, 239.3189 ,
239.45247],
[247.96744, 248.52899, 246.81934, ..., 239.25084, 239.37402,
239.52858],
[248.12334, 248.68503, 246.69528, ..., 239.22287, 239.34776,
239.5023 ]]], dtype=float32)
Coordinates:
* band (band) int64 1
* x (x) float64 3.044e+05 3.044e+05 ... 3.143e+05 3.143e+05
* y (y) float64 1.459e+06 1.459e+06 ... 1.441e+06 1.441e+06
spatial_ref int64 0
Attributes:
AREA_OR_POINT: Area
scale_factor: 1.0
add_offset: 0.0
long_name: HILLSHADE
constellation: CUSTOM
constellation_id: CUSTOM
product_path: /home/data/DS3/CI/eoreader/others/20200310T030415_WV02...
product_name: 20200310T030415_WV02_Ortho
product_filename: 20200310T030415_WV02_Ortho_BGRN_STK
instrument: CUSTOM
product_type: Ortho
acquisition_date: 20221010T141955
condensed_name: 20221010T141954_CUSTOM_Ortho
orbit_direction: None# SAR
sar_prod = reader.open(
sar_path,
custom=True,
sensor_type=SensorType.SAR,
name="20210827T162210_ICEYE_SC_GRD",
acquisition_datetime="20210827T162210",
platform="ICEYE",
product_type="GRD",
default_resolution=6.0,
band_map={VV: 1, VV_DSPK: 2},
)
2022-10-10 14:19:57,033 - [WARNING] - acquisition_datetime is not taken into account as it doesn't belong to the handled keys: ['name', 'sensor_type', 'datetime', 'band_map', 'constellation', 'instrument', 'resolution', 'product_type', 'sun_azimuth', 'sun_zenith', 'orbit_direction', 'cloud_cover']
2022-10-10 14:19:57,034 - [WARNING] - platform is not taken into account as it doesn't belong to the handled keys: ['name', 'sensor_type', 'datetime', 'band_map', 'constellation', 'instrument', 'resolution', 'product_type', 'sun_azimuth', 'sun_zenith', 'orbit_direction', 'cloud_cover']
2022-10-10 14:19:57,035 - [WARNING] - default_resolution is not taken into account as it doesn't belong to the handled keys: ['name', 'sensor_type', 'datetime', 'band_map', 'constellation', 'instrument', 'resolution', 'product_type', 'sun_azimuth', 'sun_zenith', 'orbit_direction', 'cloud_cover']
from pprint import pprint
from eoreader import utils
# Read and display metadata
mtd, _ = sar_prod.read_mtd()
pprint(utils.quick_xml_to_dict(mtd))
('custom_metadata',
{'band_map': "{'VV': 1, 'VV_DSPK': 2}",
'cloud_cover': 'None',
'constellation': 'CUSTOM',
'datetime': '2022-10-10T14:19:57.035801',
'instrument': 'CUSTOM',
'name': '20210827T162210_ICEYE_SC_GRD',
'orbit_direction': 'None',
'product_type': 'GRD',
'resolution': '47.995955510616774',
'sensor_type': 'SAR',
'sun_azimuth': 'None',
'sun_zenith': 'None'})
