Main features
Contents
Main features#
These features can be seen in the basic tutorial.
Open#
The reader singleton is your unique entry. It will create for you the product object corresponding to your satellite data.
You can load products from the cloud, see
this tutorial.
S3 and S3 Compatible Storage are working and maybe Google and Azure if rasterio supports it,
but they have not been tested.
>>> import os
>>> from reader import Reader
>>> # Path to your satellite data, ie. Sentinel-2
>>> path = r'S2A_MSIL1C_20200824T110631_N0209_R137_T30TTK_20200824T150432.zip' # You can work with the archive for S2 data
>>> # Path to your output directory (if not set, it will work in a temp directory)
>>> output = os.path.abspath('.')
>>> # Create the reader singleton
>>> reader = Reader()
>>> prod = reader.open(path, output_path=output, remove_tmp=True)
>>> # remove_tmp allows you to automatically delete processing files
>>> # such as cleaned or orthorectified bands when the product is deleted
>>> # False by default to speed up the computation if you want to use the same product in several part of your code
>>> # NOTE: you can set the output directory after the creation, that allows you to use the product condensed name
>>> prod.output = os.path.join(output, prod.condensed_name) # It will automatically create it if needed
Recognized paths#
EOReader always uses the directory containing the product files. Hereunder are the paths meant to be given to the reader.
Optical#
Sensor group |
Folder to link |
|---|---|
|
Main directory, |
|
Main directory containing the |
|
Main directory extracted or archived if Collection 2 ( |
|
Directory containing the |
|
Directory containing the |
|
Directory containing the |
SAR#
Sensor group |
Folder to link |
|---|---|
|
SAFE directory containing the |
|
Directory containing the |
|
Main directory containing the |
|
Directory containing the |
|
Directory containing the |
|
Directory containing the |
Load#
load is the function for accessing product-related bands.
It can load satellite bands, index, DEM bands and cloud bands according to this workflow:
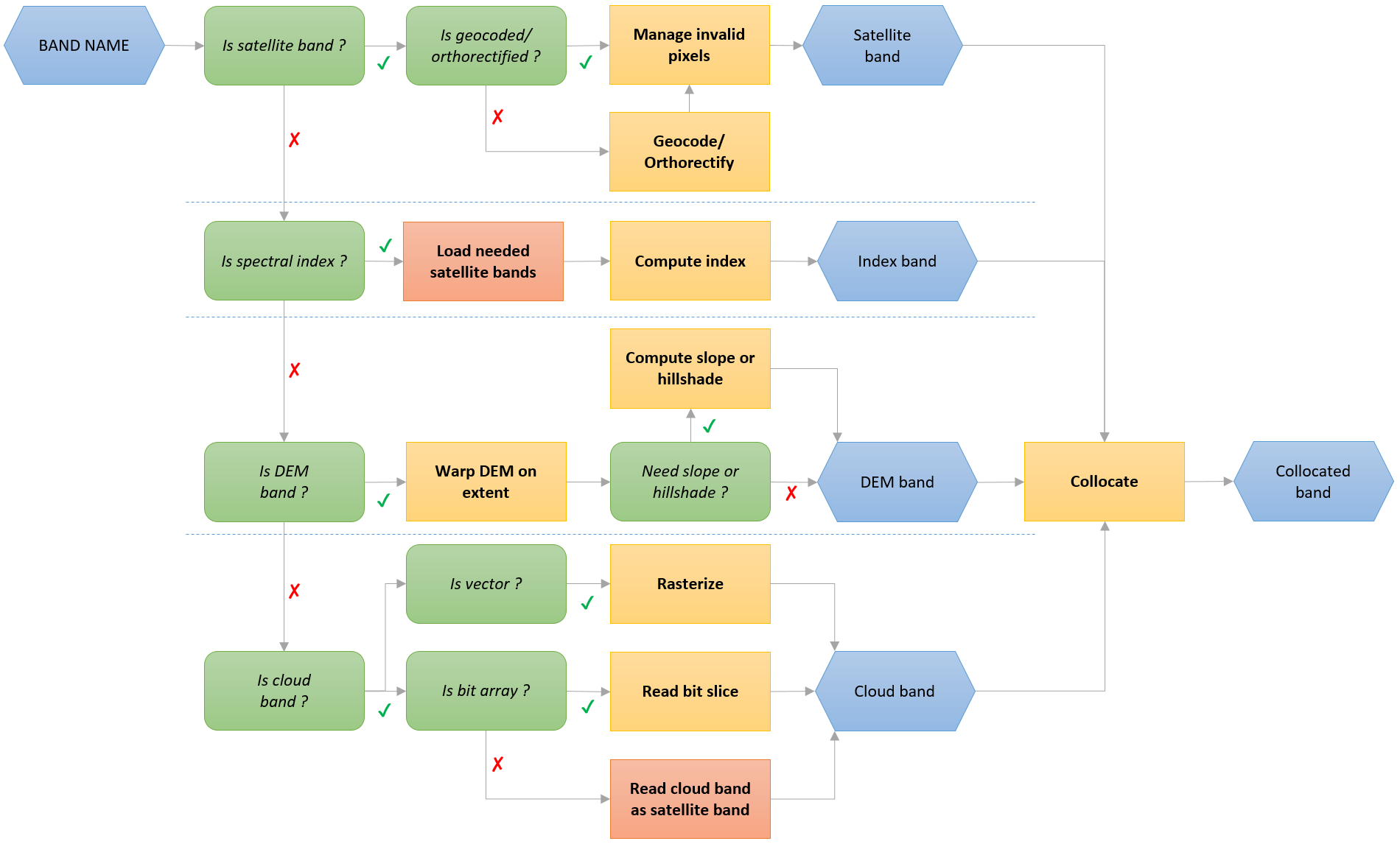
>>> import os
>>> from eoreader.reader import Reader
>>> from eoreader.bands import *
>>> import os
>>> from eoreader.env_vars import DEM_PATH
>>> path = r"S2A_MSIL1C_20200824T110631_N0209_R137_T30TTK_20200824T150432.zip"
>>> output = os.path.abspath("./output")
>>> # WARNING: you can leave the output_path empty, but EOReader will create a temporary output directory
>>> # and you won't be able to retrieve what's has been written on disk
>>> prod = Reader().open(path, output_path=output)
>>> # Specify a DEM to load DEM bands
>>> os.environ[DEM_PATH] = r"my_dem.tif"
>>> # Get the wanted bands and check if the product can produce them
>>> band_list = [GREEN, NDVI, TIR_1, SHADOWS, HILLSHADE]
>>> ok_bands = [band for band in band_list if prod.has_band(band)]
[GREEN, NDVI, HILLSHADE]
>>> # Sentinel-2 cannot produce satellite band TIR_1 and cloud band SHADOWS
>>> # Load bands
>>> bands = prod.load(ok_bands, resolution=20.) # if resolution is not specified -> load at default resolution (10.0 m for S2 data)
>>> # NOTE: every array that comes out `load` are collocated, which isn't the case if you load arrays separately
>>> # (important for DEM data as they may have different grids)
>>> bands
{<function NDVI at 0x000001C47FF05E18>: <xarray.DataArray 'NDVI' (band: 1, y: 5490, x: 5490)>
array([[[0.94786006, 0.92717856, 0.92240528, ..., 1.73572724,
1.55314477, 1.63242706],
[1.04147187, 0.93668633, 0.91499688, ..., 1.59941784,
1.52895995, 1.51386761],
[2.86996677, 1.69360304, 1.2413562 , ..., 1.61172353,
1.55742907, 1.50568275],
...,
[1.45807257, 1.61071344, 1.64620751, ..., 1.25498441,
1.42998927, 1.70447076],
[1.57802352, 1.77086658, 1.69901482, ..., 1.19999853,
1.27813254, 1.52287237],
[1.63569594, 1.66751277, 1.63474646, ..., 1.27617084,
1.22456033, 1.27022877]]])
Coordinates:
* x (x) float64 2e+05 2e+05 2e+05 ... 3.097e+05 3.098e+05 3.098e+05
* y (y) float64 4.5e+06 4.5e+06 4.5e+06 ... 4.39e+06 4.39e+06
* band (band) int32 1
spatial_ref int32 0,
<OpticalBandNames.GREEN: 'GREEN'>: <xarray.DataArray 'T30TTK_20200824T110631_B03' (band: 1, y: 5490, x: 5490)>
array([[[0.06146327, 0.06141786, 0.06100179, ..., 0.11880179,
0.12087143, 0.11468571],
[0.06123214, 0.06071094, 0.06029063, ..., 0.11465781,
0.11858906, 0.11703929],
[0.06494643, 0.06226562, 0.06169219, ..., 0.11174062,
0.11434844, 0.11491964],
...,
[0.1478125 , 0.13953906, 0.13751719, ..., 0.15949688,
0.14200781, 0.12982321],
[0.14091429, 0.12959531, 0.13144844, ..., 0.17246719,
0.156175 , 0.13453036],
[0.13521429, 0.13274286, 0.13084821, ..., 0.16064821,
0.16847143, 0.16009592]]])
Coordinates:
* x (x) float64 2e+05 2e+05 2e+05 ... 3.097e+05 3.098e+05 3.098e+05
* y (y) float64 4.5e+06 4.5e+06 4.5e+06 ... 4.39e+06 4.39e+06
* band (band) int32 1
spatial_ref int32 0,
<DemBandNames.HILLSHADE: 'HILLSHADE'>: <xarray.DataArray '20200824T110631_S2_T30TTK_L1C_150432_HILLSHADE' (band: 1, y: 5490, x: 5490)>
array([[[220., 221., 221., ..., 210., 210., 210.],
[222., 222., 221., ..., 210., 210., 210.],
[221., 221., 220., ..., 210., 210., 210.],
...,
[215., 214., 212., ..., 207., 207., 207.],
[214., 212., 211., ..., 206., 205., 205.],
[213., 211., 209., ..., 205., 204., 205.]]])
Coordinates:
* band (band) int32 1
* y (y) float64 4.5e+06 4.5e+06 4.5e+06 ... 4.39e+06 4.39e+06
* x (x) float64 2e+05 2e+05 2e+05 ... 3.097e+05 3.098e+05 3.098e+05
spatial_ref int32 0
Attributes:
grid_mapping: spatial_ref
original_dtype: uint8}
Note
Index and bands are opened as xarrays
with rioxarray, in float with the nodata set to np.nan.
The nodata written back on disk is by convention:
-9999for optical bands (saved infloat32)65535for optical bands (saved inuint16)0for SAR bands (saved infloat32), to be compliant with SNAP default nodata255for masks (saved inuint8)
For optical bands, only the pixels outside of the detector are set to nodata by default but this can be changed according to the user’s needs (see below).
Some additional arguments can be passed to this function, please see keywords for the list.
Methods to clean optical bands are best described here,
Sentinel-3 additional keywords use is highlighted in the corresponding notebook.
Stack#
stack is the function stacking all possible bands.
It is based on the load function and then just stacks the bands and write it on disk if needed.
The bands are ordered as asked in the stack.
However, they cannot be duplicated (the stack cannot contain 2 RED bands for instance)!
If the same band is asked several time, its order will be the one of the last demand.
>>> # Create a stack with the previous OK bands
>>> stack = prod.stack(ok_bands, resolution=300., stack_path=os.path.join(prod.output, "stack.tif")
<xarray.DataArray 'GREEN_NDVI_HILLSHADE' (z: 3, y: 5490, x: 5490)>
array([[[9.47860062e-01, 9.27178562e-01, 9.22405303e-01, ...,
1.73572719e+00, 1.55314481e+00, 1.63242710e+00],
[1.04147184e+00, 9.36686337e-01, 9.14996862e-01, ...,
1.59941781e+00, 1.52895999e+00, 1.51386762e+00],
[2.86996675e+00, 1.69360304e+00, 1.24135625e+00, ...,
1.61172354e+00, 1.55742908e+00, 1.50568271e+00],
...,
[1.45807254e+00, 1.61071348e+00, 1.64620745e+00, ...,
1.25498438e+00, 1.42998922e+00, 1.70447075e+00],
[1.57802355e+00, 1.77086663e+00, 1.69901478e+00, ...,
1.19999850e+00, 1.27813256e+00, 1.52287233e+00],
[1.63569593e+00, 1.66751277e+00, 1.63474643e+00, ...,
1.27617085e+00, 1.22456038e+00, 1.27022874e+00]],
[[6.14632666e-02, 6.14178553e-02, 6.10017851e-02, ...,
1.18801787e-01, 1.20871432e-01, 1.14685714e-01],
[6.12321422e-02, 6.07109368e-02, 6.02906235e-02, ...,
1.14657812e-01, 1.18589066e-01, 1.17039286e-01],
[6.49464279e-02, 6.22656234e-02, 6.16921857e-02, ...,
1.11740626e-01, 1.14348434e-01, 1.14919640e-01],
[1.47812501e-01, 1.39539063e-01, 1.37517184e-01, ...,
1.59496874e-01, 1.42007813e-01, 1.29823208e-01],
[1.40914291e-01, 1.29595309e-01, 1.31448433e-01, ...,
1.72467187e-01, 1.56175002e-01, 1.34530351e-01],
[1.35214284e-01, 1.32742852e-01, 1.30848214e-01, ...,
1.60648212e-01, 1.68471426e-01, 1.60095915e-01]],
[[2.20000000e+02, 2.21000000e+02, 2.21000000e+02, ...,
2.10000000e+02, 2.10000000e+02, 2.10000000e+02],
[2.22000000e+02, 2.22000000e+02, 2.21000000e+02, ...,
2.10000000e+02, 2.10000000e+02, 2.10000000e+02],
[2.21000000e+02, 2.21000000e+02, 2.20000000e+02, ...,
2.10000000e+02, 2.10000000e+02, 2.10000000e+02],
...,
[2.15000000e+02, 2.14000000e+02, 2.12000000e+02, ...,
2.07000000e+02, 2.07000000e+02, 2.07000000e+02],
[2.14000000e+02, 2.12000000e+02, 2.11000000e+02, ...,
2.06000000e+02, 2.05000000e+02, 2.05000000e+02],
[2.13000000e+02, 2.11000000e+02, 2.09000000e+02, ...,
2.05000000e+02, 2.04000000e+02, 2.05000000e+02]]], dtype=float32)
Coordinates:
* x (x) float64 2e+05 2e+05 2e+05 ... 3.097e+05 3.098e+05 3.098e+05
* y (y) float64 4.5e+06 4.5e+06 4.5e+06 ... 4.39e+06 4.39e+06
spatial_ref int32 0
* z (z) MultiIndex
- variable (z) object 'GREEN' 'NDVI' 'HILLSHADE'
- band (z) int64 1 1 1
Attributes:
long_name: ['GREEN', 'NDVI', 'HILLSHADE']
Some additional arguments can be passed to this function, please see keywords for the list.
Methods to clean optical bands are best described here,
Sentinel-3 additional keywords use is highlighted in the corresponding notebook.
Read Metadata#
EOReader gives you the access to the metadata of your product as a lxml.etree._Element followed by the namespace you may need to read them
>>> # Access the raw metadata as an lxml.etree._Element and its namespaces as a dict:
>>> mtd, nmsp = prod.read_mtd()
(
<Element {https://psd-14.sentinel2.eo.esa.int/PSD/S2_PDI_Level-1C_Tile_Metadata.xsd}Level-1C_Tile_ID at 0x1e396036ec8>,
{'n1': '{https://psd-14.sentinel2.eo.esa.int/PSD/S2_PDI_Level-1C_Tile_Metadata.xsd}'}
)
>>> # You can access a field like that:
>>> datastrip_id = mtd.findtext(".//DATASTRIP_ID")
>>> # Pay attention, for some products you will need a namespace, i.e. for planet data:
>>> # name = mtd.findtext(f".//{nsmap['eop']}identifier")
Note
Landsat Collection 1 have no metadata with XML format, so the XML is simulated from the text file.
Note
Sentinel-3 sensors have no metadata file but have global attributes repeated in every NetCDF files. This is what you will have when calling this function:
absolute_orbit_numbercommentcontactcreation_timehistoryinstitutionnetCDF_versionproduct_namereferencesresolutionsourcestart_offsetstart_timestop_timetitleac_subsampling_factor(OLCIonly)al_subsampling_factor(OLCIonly)track_offset(SLSTRonly)
Plot#
If a quicklook exists, the user can plot the product. Always existing for VHR and SAR data, more rarely for other optical sensors. See Optical and SAR tutorials for examples.
>>> # Plot product
>>> prod.plot()
Other features#
CRS#
Get the product CRS, always in UTM
>>> # Product CRS (always in UTM)
>>> prod.crs()
CRS.from_epsg(32630)
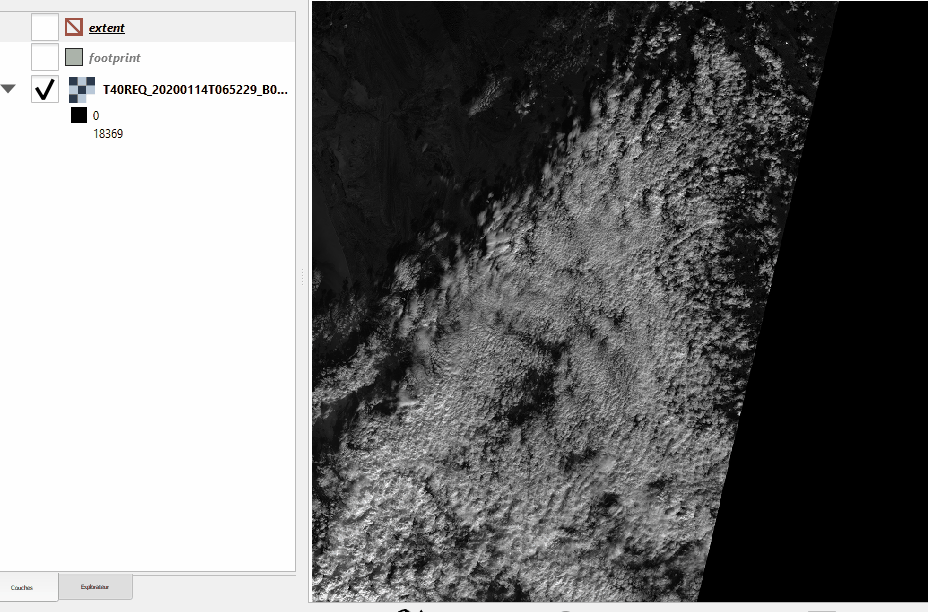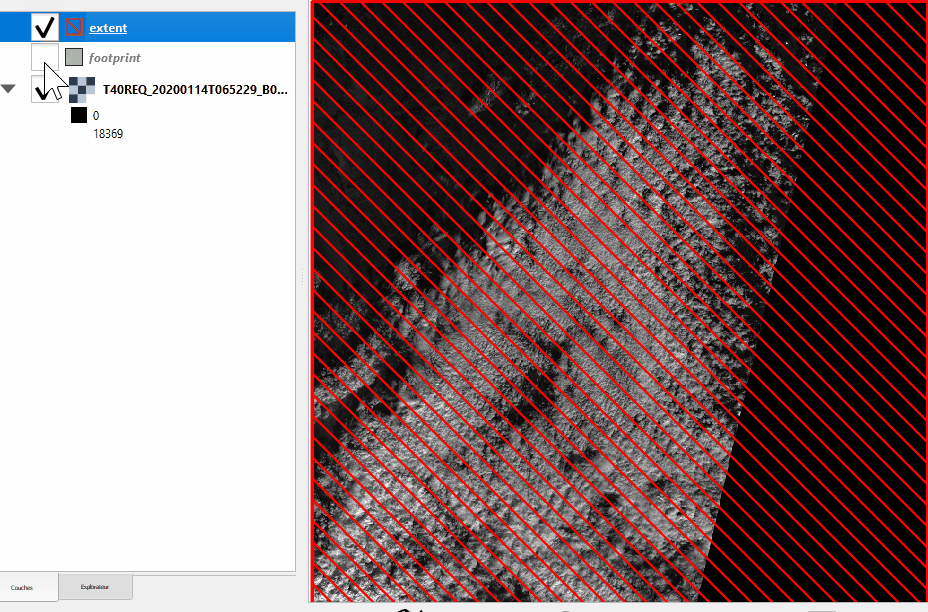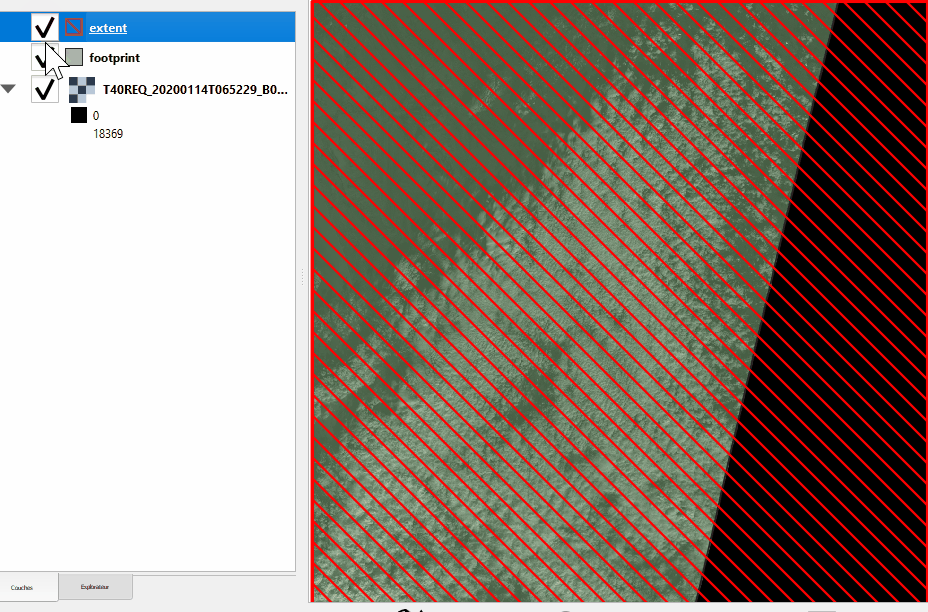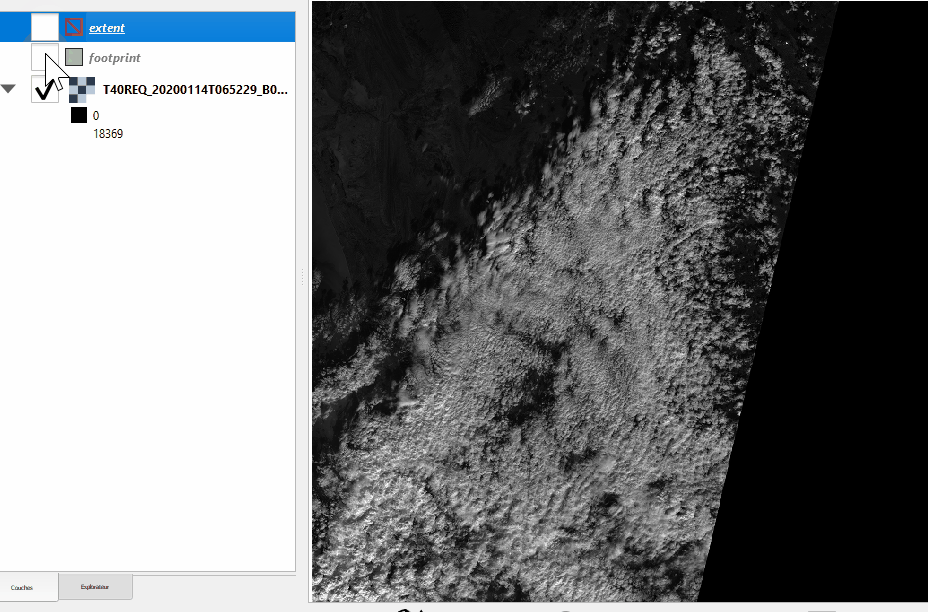
Extent and footprint#
Get the product extent and footprint, always in UTM as a gpd.GeoDataFrame
>>> # Full extent of the bands as a geopandas GeoDataFrame
>>> prod.extent()
geometry
0 POLYGON((309780.000 4390200.000, 309780.000 4...
>>> # Footprint: extent of the useful pixels (minus nodata) as a geopandas GeoDataFrame
>>> prod.footprint()
geometry
0 POLYGON Z((199980.000 4390200.000 0.000, 1999...
Please note the difference between footprint and extent:
Without nodata |
With nodata |
|---|---|
|
|
Solar angles#
Get optical product azimuth (between [0, 360] degrees) and zenith solar angles, useful for computing the Hillshade for example.
>> > # Get azimuth and zenith solar angles
>> > prod.get_mean_sun_angles()
(151.750970396115, 35.4971906983449)
Cloud Cover#
Get optical product cloud cover as specified in the metadata
>> > # Get cloud cover
>> > prod.get_cloud_cover()
55.5
Orbit direction#
Get product optical direction (useful especially for SAR data), as a OrbitDirection (ASCENDING or DESCENDING).
Always specified in the metadata for SAR sensors, set to DESCENDING by default for optical data if not existing.
>> > # Get orbit direction
>> > prod.get_orbit_direction()
< OrbitDirection.DESCENDING: 'DESCENDING' >